Introduction to Flexible Clustering of (Mixed-With-)Ordinal Data
Source:vignettes/Intro2Flexord.Rmd
Intro2Flexord.RmdPackage description and contents
Package flexord is an add on-package to packages flexclust and flexmix that provide suites for partitioning and model-based clustering with flexible method switching and comparison.
We provide additional distance and centroid calculation functions, and additional model drivers for component distributions that are tailored towards ordinal, or mixed-with-ordinal data. These new methods can easily be plugged into the capabilities for clustering provided by flexclust and flexmix.
By plugging them into the flex-scheme, they can be used for:
- one-off K-centroids and model-based clustering (via
flexclust::kccaandflexmix::flexmix), - repeated clustering runs with various cluster numbers
k(viaflexclust::stepFlexclustandflexmix::stepFlexmix), - bootstrapping repeated clustering runs with various cluster numbers
kfor K-centroids clustering (viaflexclust::bootFlexclust), - applying the various methods for the resulting objects, such as
predict,plot,barchart, …
| Clustering Type | Function Type | Function Name | Method | Scale Assumptions | NA Handling | Source |
|---|---|---|---|---|---|---|
| Partitioning (K-centroids) | distance |
distSimMatch
|
Simple Matching Distance | nominal | not implemented | Kaufman and Rousseeuw (1990), p. 19 |
distGDM2
|
GDM2 distance for ordinal data | ordinal | not implemented | Walesiak and Dudek (2010); Ernst et al. (2025) | ||
distGower
|
Gower’s distance | mixed-with-ordinal | upweighing of present variables | Kaufman and Rousseeuw (1990), p. 32-37 | ||
| centroid |
centMode
|
Mode as centroid | nominal | not implemented | Weihs et al. (2005); Leisch (2006) | |
centMin
|
Factor level with minimal distance as centroid | nominal/ordinal | not implemented | Ernst et al. (2025) | ||
centOptimNA
|
Centroid calculation by general purpose optimizer | numeric | complete-case analysis | Leisch (2006) | ||
| wrapper |
kccaExtendedFamily
|
Creates a kccaFamily object pre-configured for kModes-,
kGDM2- or kGower clustering
|
||||
| Model-based | driver |
FLXMCregnorm
|
Regularized multivariate normal distribution | numeric | not implemented | Fraley and Raftery (2007); Ernst et al. (2025) |
FLXMCregmultinom
|
Regularized multivariate multinomial distribution | nominal | not implemented | Galindo Garre and Vermunt (2006); Ernst et al. (2025) | ||
FLXMCregbinom
|
Regularized multivariate binomial distribution | ordinal | not implemented | Ernst et al. (2025) | ||
FLXMCbetabinomial
|
Regularized multivariate beta-binomial distribution | ordinal | not implemented | Kondofersky (2008); Ernst et al. (2025) |
Example 1: Clustering purely nominal data
We load necessary packages and set a random seed for reproducibility.
As an example for purely nominal data, we will use the classic
Titanic data set:
titanic_df <- data.frame(Titanic)
titanic_df <- titanic_df[rep(1:nrow(titanic_df), titanic_df$Freq), -5]
str(titanic_df)
#> 'data.frame': 2201 obs. of 4 variables:
#> $ Class : Factor w/ 4 levels "1st","2nd","3rd",..: 3 3 3 3 3 3 3 3 3 3 ...
#> $ Sex : Factor w/ 2 levels "Male","Female": 1 1 1 1 1 1 1 1 1 1 ...
#> $ Age : Factor w/ 2 levels "Child","Adult": 1 1 1 1 1 1 1 1 1 1 ...
#> $ Survived: Factor w/ 2 levels "No","Yes": 1 1 1 1 1 1 1 1 1 1 ...Partitioning approach
We can conduct K-centroids clustering with the kModes algorithm directly on the data frame1:
kcca(titanic_df, k = 4, family = kccaExtendedFamily('kModes'))
#> kcca object of family 'kModes'
#>
#> call:
#> kcca(x = titanic_df, k = 4, family = kccaExtendedFamily("kModes"))
#>
#> cluster sizes:
#>
#> 1 2 3 4
#> 140 396 287 1378Let us assume that for some reason we are unhappy with the mode as a centroid, and rather want to use an optimized centroid value, by choosing the factor level for which Simple Matching distance2 is minimal:
kcca(titanic_df, k = 4,
family = kccaFamily(dist = distSimMatch,
cent = \(y) centMin(y, dist = distSimMatch,
xrange = 'columnwise')))
#> kcca object of family 'distSimMatch'
#>
#> call:
#> kcca(x = titanic_df, k = 4, family = kccaFamily(dist = distSimMatch,
#> cent = function(y) centMin(y, dist = distSimMatch, xrange = "columnwise")))
#>
#> cluster sizes:
#>
#> 1 2 3 4
#> 369 737 317 778This already showcases one of the advantages of package flexclust: As the name suggests, we are quickly able to mix and match our distance and centroid functions, and quickly create our own K-centroids algorithms.
Furthermore, flexclust allows us to decrease the
influence of randomness via running the algorithm several times, and
keeping only the solution with the minimum within cluster distance. This
can be done for one specific number of clusters k or
several values k:
stepFlexclust(data.matrix(titanic_df), k = 2:4, nrep = 3,
family = kccaExtendedFamily('kModes'))
#> 2 : * * *
#> 3 : * * *
#> 4 : * * *
#> stepFlexclust object of family 'kModes'
#>
#> call:
#> stepFlexclust(x = data.matrix(titanic_df), k = 2:4, nrep = 3,
#> family = kccaExtendedFamily("kModes"))
#>
#> iter converged distsum
#> 1 NA NA 651.50
#> 2 200 FALSE 413.50
#> 3 200 FALSE 278.75
#> 4 200 FALSE 246.00The output above shows the solutions with lowest within cluster distance out of 3 runs for 2 to 4 clusters, in comparison to 1 big cluster. However, none of the algorithms converged. Presumably this is due to observations which have the same distance to two centroids and which are randomly assigned to one of the two centroids, implying that the partitions are still changing in each iteration, even if the centroids do not change.
Selecting a suitable number of clusters based on the output of
stepFlexclust might be still difficult. This is where
bootFlexclust comes in. In bootFlexclust,
nboot bootstrap samples of the original data are drawn, on
which stepFlexclust is performed for each k.
This results in
knboot
best out of nrep clustering solutions obtained for each
bootstrap data set. Based on these solutions cluster memberships are
predicted for the original data set, and the stability of these
partitions is tested via the Adjusted Rand Index (Hubert and Arabie 1985):
(nom <- bootFlexclust(data.matrix(titanic_df),
k = 2:4, nrep = 3, nboot = 5,
family = kccaExtendedFamily('kModes')))
#> An object of class 'bootFlexclust'
#>
#> Call:
#> bootFlexclust(x = data.matrix(titanic_df), k = 2:4, nboot = 5,
#> nrep = 3, family = kccaExtendedFamily("kModes"))
#>
#> Number of bootstrap pairs: 5Note that ridiculously few repetitions are used for the sake of having a short run time.
The resulting ARIs can be quickly visualized via a predefined plotting method:
plot(nom)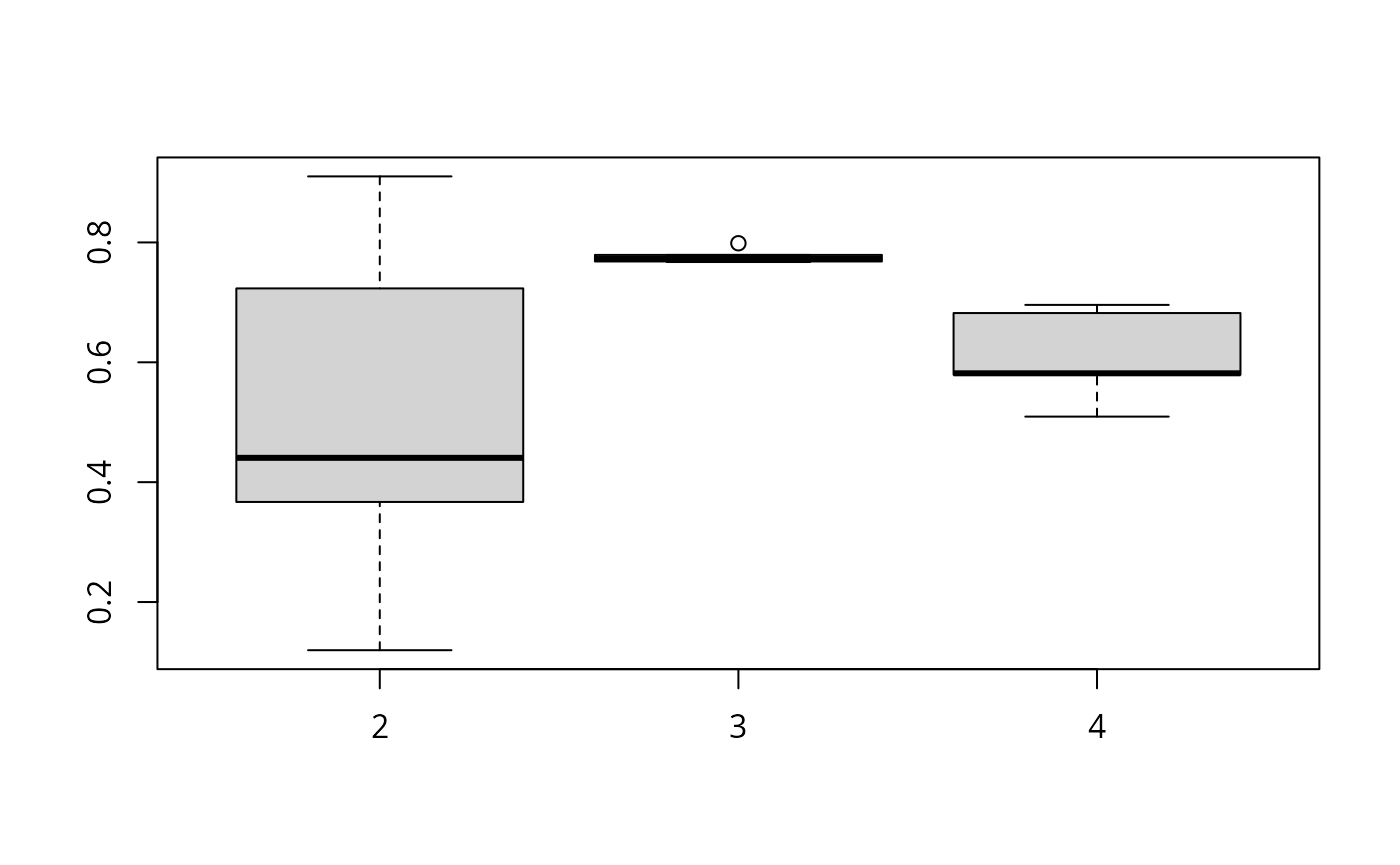
Our plot indicates that out of the 2 to 4 cluster solutions, a three cluster solution has the highest median ARI out of 5 runs.
Now, after deciding on a suitable cluster number, we could select the
corresponding cluster solution from kcca or
stepFlexclust, and make use of the further visualization,
prediction, and other tools. For this, we refer to the documentation
available in Leisch (2006) and Dolnicar, Grün, and Leisch (2018).
Model-based approach
We also offer an algorithm specifically designed for model-based
clustering of unordered categorical data via a regularized multinomial
distribution. The multinomial driver also supports varying number of
categories between variables. Then we call flexmix with a
model driver specifying the number of categories:
titanic_ncats = apply(data.matrix(titanic_df), 2, max)
flexmix(formula = data.matrix(titanic_df) ~ 1, k = 3,
model = FLXMCregmultinom(r = titanic_ncats))
#>
#> Call:
#> flexmix(formula = data.matrix(titanic_df) ~ 1, k = 3, model = FLXMCregmultinom(r = titanic_ncats))
#>
#> Cluster sizes:
#> 1 2 3
#> 1208 364 629
#>
#> convergence after 125 iterationsAs we have to estimate many category probabilities across multiple clusters some of those may become numerically zero. To avoid this we may use the regularization parameter , which acts if we added observations according to the population mean to each component:
flexmix(data.matrix(titanic_df) ~ 1, k = 3,
model=FLXMCregmultinom(r = titanic_ncats, alpha = 1))
#>
#> Call:
#> flexmix(formula = data.matrix(titanic_df) ~ 1, k = 3, model = FLXMCregmultinom(r = titanic_ncats,
#> alpha = 1))
#>
#> Cluster sizes:
#> 1 2 3
#> 364 629 1208
#>
#> convergence after 56 iterationsflexmix also offers a step method, where
the EM algorithm for each k is restarted nrep
times, and only the maximum likelihood solution is retained:
(nom <- stepFlexmix(data.matrix(titanic_df)~1, k = 2:4,
nrep = 3, #please increase for real-life use
model = FLXMCregmultinom(r = titanic_ncats)))
#> 2 : * * *
#> 3 : * * *
#> 4 : * * *
#>
#> Call:
#> stepFlexmix(data.matrix(titanic_df) ~ 1, model = FLXMCregmultinom(r = titanic_ncats),
#> k = 2:4, nrep = 3)
#>
#> iter converged k k0 logLik AIC BIC ICL
#> 2 38 TRUE 2 2 -5327.340 10680.68 10754.74 10986.90
#> 3 103 TRUE 3 3 -5202.816 10445.63 10559.57 11036.09
#> 4 121 TRUE 4 4 -5176.038 10406.08 10559.89 11454.98The output of this is the best out of three clusterings for 3
different values of k. We are also already provided with
different model selection criteria, namely, AIC, BIC and ICL.
Similar to package flexclust in the partitioning case, package flexmix also offers various plotting methods for the returned objects. We just showcase one here for simplicity:
plot(nom)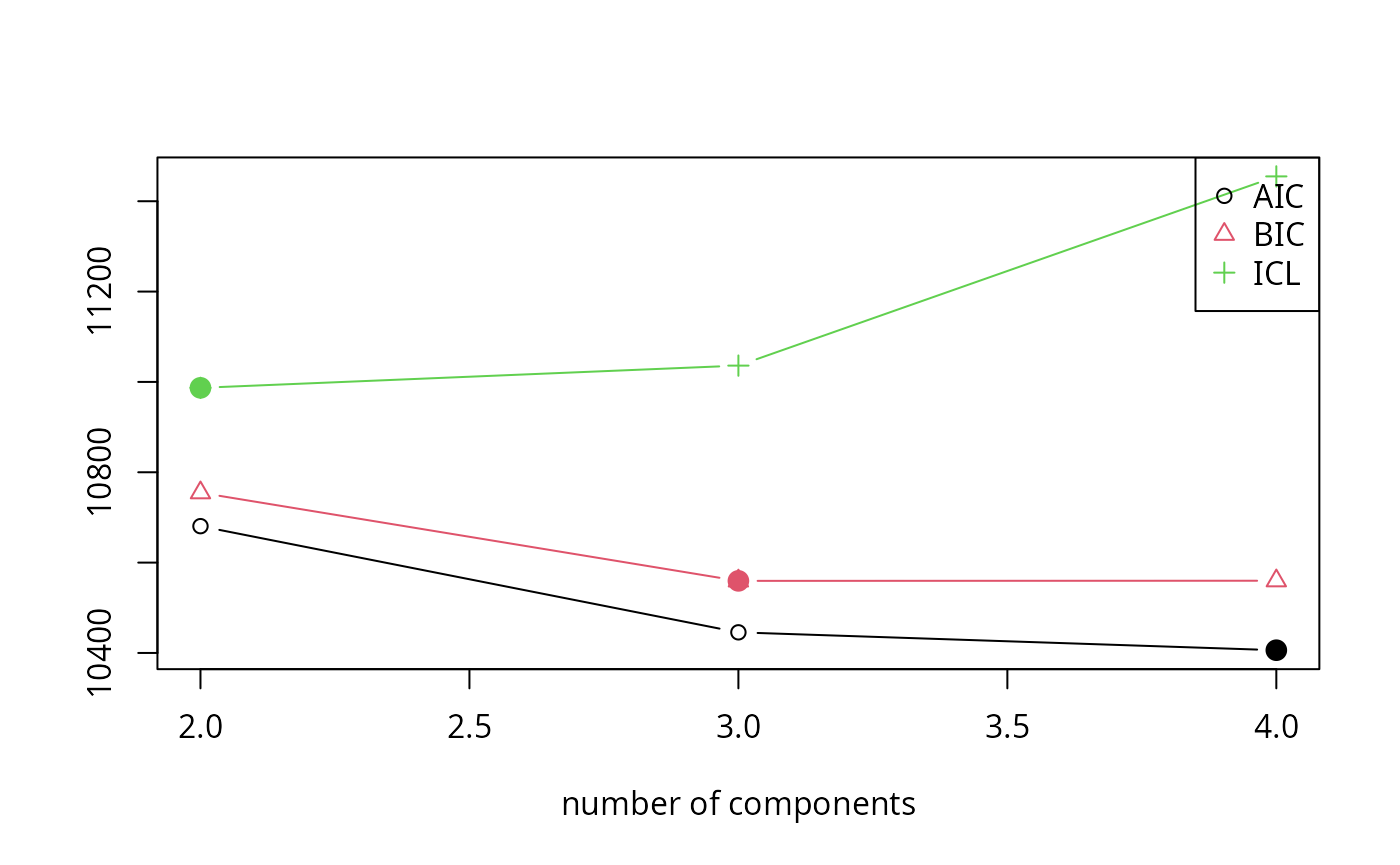
For more information on the further methods and utilities offered,
check out the documentation for flexmix
(browseVignettes('flexmix')).
Example 2: Clustering purely ordinal data
Our next example data set is from a survey conducted among 563 Australians in 2015 where they indicated on a scale from 1-5 how inclined they are to take 6 types of risks. It consists of purely ordinal variables without missing values, and the response level length is the same for all variables.
data("risk", package = "flexord")
str(risk)
#> int [1:563, 1:6] 3 1 2 1 5 5 1 5 1 3 ...
#> - attr(*, "dimnames")=List of 2
#> ..$ : NULL
#> ..$ : chr [1:6] "Recreational" "Health" "Career" "Financial" ...
colnames(risk)
#> [1] "Recreational" "Health" "Career" "Financial" "Safety"
#> [6] "Social"Partitioning approach
In our package, we offer two partitioning methods designed for
ordinal data: Firstly, we can apply Gower’s distance from
distGower to purely ordinal data, which results in using
Manhattan distance (as provided also in
flexclust::distManhattan) with previous scaling as
described by Kaufman and Rousseeuw (1990)
and Gower’s upweighing of non-missing values:
kcca(risk, k = 4, family = kccaExtendedFamily('kGower'))
#> kcca object of family 'kGower'
#>
#> call:
#> kcca(x = risk, k = 4, family = kccaExtendedFamily("kGower"))
#>
#> cluster sizes:
#>
#> 1 2 3 4
#> 133 123 106 201The default centroid for this family is the general purpose optimizer
centOptimNA, which is the general purpose optimizer
flexclust::centOptim, just with NA removal. In our case of
purely ordinal data with no missing values, we could also choose the
median as a centroid:
kcca(risk, k = 4,
family = kccaExtendedFamily('kGower', cent = centMedian))
#> kcca object of family 'kGower'
#>
#> call:
#> kcca(x = risk, k = 4, family = kccaExtendedFamily("kGower", cent = centMedian))
#>
#> cluster sizes:
#>
#> 1 2 3 4
#> 300 65 90 108This results in kMedians with previous scaling, and non-missing value
upweighing3. In our risk example with no
NAs and equal level lengths for all variables,
flexclust::kccaFamily('kmedians') would suffice, but there
are still many data situations where the kGower approach
will be preferable.
As a second alternative designed specifically for ordinal data without missing values, we implement the GDM2 distance for ordinal data by Walesiak and Dudek (2010), which conduct only relational operations on ordinal variables. We have reformulated it for use in K-centroids analysis in Ernst et al. (2025), and implemented it in the package:
kcca(risk, k = 3, family = kccaExtendedFamily('kGDM2'))
#> kcca object of family 'kGDM2'
#>
#> call:
#> kcca(x = risk, k = 3, family = kccaExtendedFamily("kGDM2"))
#>
#> cluster sizes:
#>
#> 1 2 3
#> 101 77 385Same as in kGower, a default general optimizer centroid
is applied, which we could replace as desired.
Another parameter used in both kGower and
kGDM2 is xrange. Both algorithms require
information on the range of the data object for data preprocessing, one
for scaling, the other for transforming the data to empirical
distributions. The range calculation can be influenced in the following
ways: We can use the range of the whole x (argument
all, the default for kGDM2), columnwise ranges
(xrange=columnwise), a vector specifying the range of the
data set, or a list of length ncol(x) with range vectors
for each column. Let us assume that the highest possible response to the
risk questions was Extremely often (6), but it
was never chosen by any of the respondents. We can take the new assumed
full range of the data into account:
kcca(risk, k = 3,
family = kccaExtendedFamily('kGDM2', xrange = c(1, 6)))
#> kcca object of family 'kGDM2'
#>
#> call:
#> kcca(x = risk, k = 3, family = kccaExtendedFamily("kGDM2", xrange = c(1,
#> 6)))
#>
#> cluster sizes:
#>
#> 1 2 3
#> 97 43 423Again, the distances, centroids, and wrapper alternatives presented can be used also in the further capabilities of flexclust.
Model-based approach
We also offer drivers for two distributions for ordinal data, which are the binomial distribution and its extension the beta-binomial distribution:
flexmix(risk~1, k=3, model=FLXMCregbinom(size=5))
#>
#> Call:
#> flexmix(formula = risk ~ 1, k = 3, model = FLXMCregbinom(size = 5))
#>
#> Cluster sizes:
#> 1 2 3
#> 284 28 251
#>
#> convergence after 58 iterations
flexmix(risk~1, k=3, model=FLXMCregbetabinom(size=5, alpha=1))
#>
#> Call:
#> flexmix(formula = risk ~ 1, k = 3, model = FLXMCregbetabinom(size = 5,
#> alpha = 1))
#>
#> Cluster sizes:
#> 1 2 3
#> 271 36 256
#>
#> convergence after 56 iterationsIn both cases we specify the number of trials of the binomial
distribution (size). For both distributions we can also use
a regularization parameter alpha that draws the component
estimates towards the population mean. While this incurs small
distortions it can be helpful to avoid boundary estimates.
The beta-binomial distribution is parameterized by two parameters
a and b and is therefore more flexible than
the binomial. It may potentially perform better in more difficult
clustering scenarios even if we assume the original data was drawn from
a binomial mixture (Ernst et al. (2025)).
Your mileage may vary.
We can further use the capabilities of stepFlexmix and
the corresponding plot functions. See Example 1.
Treating the data as purely nominal
Treating ordered categorical data as unordered is a frequent
approach. In fact, in our simulation study it was a quite competitive
approach for model-based methods. However, applying kmodes
to ordered data brought subpar results in the partitioning ambit (Ernst et al. 2025). For How-Tos, please look at
Example 1.
Treating the data as equidistant (=integer)
Also treating ordered categorical data as integer values is at least
as common as nominalization. In fact, some of the methods presented
above, such as kGower - as used above on purely ordinal
data without missing values - make only lax concessions towards
ordinality. Depending on data characteristics and method group applied,
this approach may also be a very good choice (Ernst et al. 2025).
We do not offer any new methods for this in the partitioning ambit, as already many options are available in flexclust.
In the model-based ambit we offer additional capabilities via
FLXMCregnorm, which, as mentioned, is a driver for
clustering with multivariate normal distributions while allowing for
regularization (same as is the case for FLXMCregmultinom,
regularization can help avoid degenerate solutions):
flexmix(risk ~ 1, k = 3,
model = FLXMCregnorm(kappa_p = 0.1, #regularization parameter
G = 3)) #number of clusters, used for scale calculation
#>
#> Call:
#> flexmix(formula = risk ~ 1, k = 3, model = FLXMCregnorm(kappa_p = 0.1,
#> G = 3))
#>
#> Cluster sizes:
#> 1 2
#> 505 58
#>
#> convergence after 43 iterationsThis implements the univariate case with different variances from
Fraley and Raftery (2007) . In the source
paper they use an inverse gamma distribution as conjugate prior. Using
the prior we can again avoid boundary estimates. Most notably the
shrinkage parameter kappa_p (the suffix _p
stands for prior), acts as if we added kappa_p observations
according to the population mean to each component.
The scale parameter zeta_p is computed by default as the
empirical variance divided by the square of the number of components. In
this case we need to pass G=3. We could however specify a
value for zeta_p and then omit the parameter G
(we can’t have both parameter at the same time, therefore
zeta_p takes precedence if both were given).
For more details see Fraley and Raftery
(2007) or package Mclust.
Again, the model can be plugged into all of the further tools offered by flexmix, for some example usages see Example 1.
Example 3: Clustering mixed-type data with missing values
data("vacmot", package = "flexclust")
vacmot2 <- cbind(vacmotdesc,
apply(vacmot, 2, as.logical))
vacmot2 <- vacmot2[, c('Gender', 'Age', 'Income2', 'Relationship.Status', 'Vacation.Behaviour',
sample(colnames(vacmot), 3, replace = FALSE))]
vacmot2$Income2 <- as.ordered(vacmot2$Income2)
str(vacmot2)
#> 'data.frame': 1000 obs. of 8 variables:
#> $ Gender : Factor w/ 2 levels "Male","Female": 2 2 1 2 1 2 1 1 2 2 ...
#> $ Age : num 25 31 21 18 61 63 58 41 36 56 ...
#> $ Income2 : Ord.factor w/ 5 levels "<30k"<"30-60k"<..: 2 5 4 2 1 2 1 2 4 2 ...
#> $ Relationship.Status : Factor w/ 5 levels "single","married",..: 1 2 1 1 2 2 3 4 3 2 ...
#> $ Vacation.Behaviour : num 2.07 2 1.23 2.17 1.72 ...
#> $ do sports : logi FALSE FALSE FALSE FALSE FALSE FALSE ...
#> $ maintain unspoilt surroundings: logi FALSE FALSE FALSE FALSE TRUE FALSE ...
#> $ luxury / be spoilt : logi FALSE TRUE FALSE TRUE FALSE FALSE ...
colMeans(is.na(vacmot2))*100
#> Gender Age
#> 0.0 0.0
#> Income2 Relationship.Status
#> 6.6 0.4
#> Vacation.Behaviour do sports
#> 2.5 0.0
#> maintain unspoilt surroundings luxury / be spoilt
#> 0.0 0.0For our last example, we chose a merged data set that is shared in
flexclust. In flexclust, it is
provided in the object vacmot as a
matrix of binary responses to questions on travel motives posed to
Australians in 2006, plus a separate data frame vacmotdesc
with 12 demographic variables for each respondent.
This data set has been thoroughly explored for clustering in the
field of market segmentation research, see for example Dolnicar and Leisch (2008). We now use it as a
data example for a mixed-data case with a moderate amount of
missingness. For this, we chose 1 symmetric binary variable (Gender,
which was collected as Male/Female in 2006), 2 numeric variables (Age
and Vacation Behaviour4), 1 unordered categorical variable
(Relationship Status), 1 ordered categorical variable (Income2, which is
a recoding of Income), and 3 randomly selected asymmmetric
binary variables (3 of the 20 questions on whether a specific travel
motive applies to a respondent). Missing values are present, but the
percentage is low5.
Partitioning approach
Currently, we only offer one method for mixed-type data with missing
values, which is kGower (scaling and distances as proposed
by Gower (1971) and Kaufman and Rousseeuw (1990), and a general
purpose optimizer centroid as provided in flexclust,
but with NA omission):
kcca(vacmot2, k = 3, family = kccaExtendedFamily('kGower'))
#> kcca object of family 'kGower'
#>
#> call:
#> kcca(x = vacmot2, k = 3, family = kccaExtendedFamily("kGower"))
#>
#> cluster sizes:
#>
#> 1 2
#> 318 682In our example above, the default methods for each variable type are used (Simple Matching Distance for the categorically coded variables, squared Euclidean distance for the numerically/integer coded variables, Manhattan distance for ordinal variables, and Jaccard distance for logically coded variables).
We could instead provide a vector of length
ncol(vacmot2) where each distance measure to be used is
specified. Let us assume that we have many outliers in the variable
Age, that we consider Vacation.Behaviour an
ordered factor as well, and that the three binary responses to vacation
motives are symmetric instead of asymmetric6, and for this reason
want to evaluate the first with Manhattan distance, and the latter with
Euclidean distance7:
colnames(vacmot2)
#> [1] "Gender" "Age"
#> [3] "Income2" "Relationship.Status"
#> [5] "Vacation.Behaviour" "do sports"
#> [7] "maintain unspoilt surroundings" "luxury / be spoilt"
xmthds <- c('distSimMatch', rep('distManhattan', 3),
'distSimMatch', rep('distEuclidean', 3))
kcca(vacmot2, k = 3,
family = kccaExtendedFamily('kGower', xmethods = xmthds))
#> kcca object of family 'kGower'
#>
#> call:
#> kcca(x = vacmot2, k = 3, family = kccaExtendedFamily("kGower",
#> xmethods = xmthds))
#>
#> cluster sizes:
#>
#> 1 2 3
#> 276 296 428For kGower, all numeric/integer and ordered variables
are scaled as proposed by Kaufman and Rousseeuw
(1990), by centering around their minimum and dividing by the
range. This means that also for kGower, the range of the
variables will influence the clustering results. Same as for
kGDM2 (Example 2), we can specify the
range to be used in parameter xrange. In the case of
kGower, the default value is columnwise, where
the range for each column is calculated separately.
Again, the distance, centroid and wrapper functions can be used in the further tools provided by flexclust, for examples on that see Example 1.
References
Internally, it will be converted to a
data.matrix. However, as only equality operations and frequency counts are used, this is of no consequence.↩︎i.e., the mean disagreement count↩︎
Note to selves: Strictly, it doesn’t, as Fritz never made his centMedianNA public↩︎
Mean environmental friendly behaviour score, ranging from 1 to 5↩︎
This is by choice. While Gower’s distance is designed to handle missingness via variable weighting, and the general optimizer used here is written to omit NAs, both methods will degenerate with high percentages of missing values. While we have not yet determined the critical limit, we have successfully run the algorithm on purely ordinal data with MCAR missingness percentages of up to 30%. However, common sense dictates that solutions obtained for such high missingness percentages need to be treated with caution.↩︎
meaning that 2 disagreements are just as important as 2 agreements↩︎
we could achieve symmetric treatment also via Simple Matching Distance↩︎